Output Files
For this example we will use the directory 01_clean_reads previously created with the clean module. We run the following Captus command to assemble our cleaned reads:
captus assemble --reads 01_clean_reads --sample_reads_target 1000000 --max_contig_gc 60.0
We are including the option --sample_reads_target 1000000 to show the expected output even though this option will not be very commonly used. Additionally, the option --max_contig_gc 60.0 is used to filter contigs with GC content over 60% after assembly, the filtered contigs are always saved to removed_contigs.fasta in case the filtering has to be repeated after changing the thresholds.
After the run is finished we should see a new directory called 02_assemblies with the following structure and files:
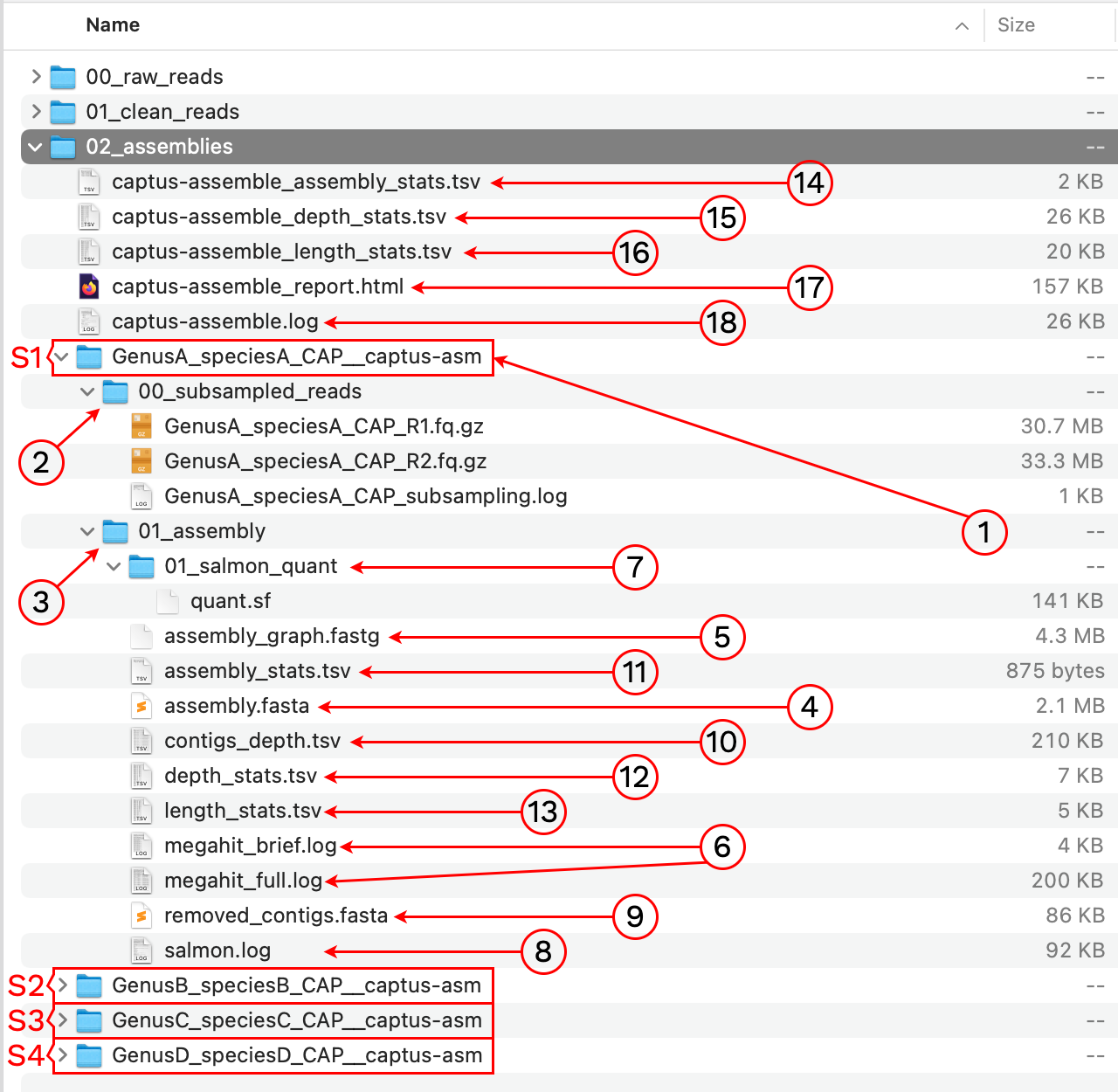
1. [sample]__captus-asm
A subdirectory ending in __captus-asm is created to contain the assembly of each sample separately (S1, S2, S3, and S4 in the image).
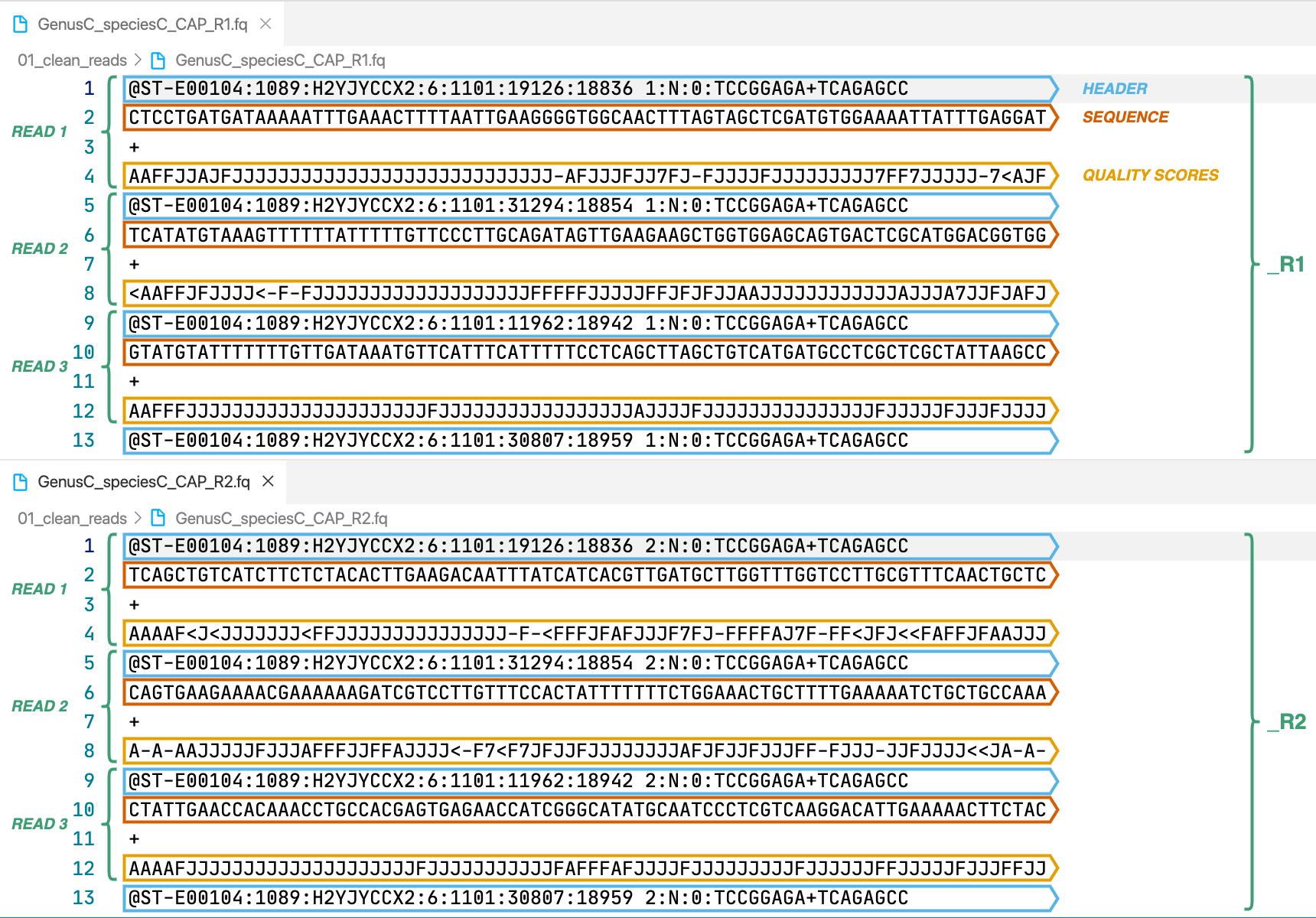
2. 00_subsampled_reads
This directory is only created when the option --sample_reads_target is used. It contains the subsampled reads in FASTQ format that were used for the assembly.
3. 01_assembly
This directory contains the FASTA and FASTG assembly files as well as assembly statistics and logs.
4. assembly.fasta
The main assembly file in FASTA format, this file contains the contigs assembled by MEGAHIT and filtered according --max_contig_gc and --min_contig_depth. The sequence headers are modified by Captus to resemble the headers produced by the assembler Spades.
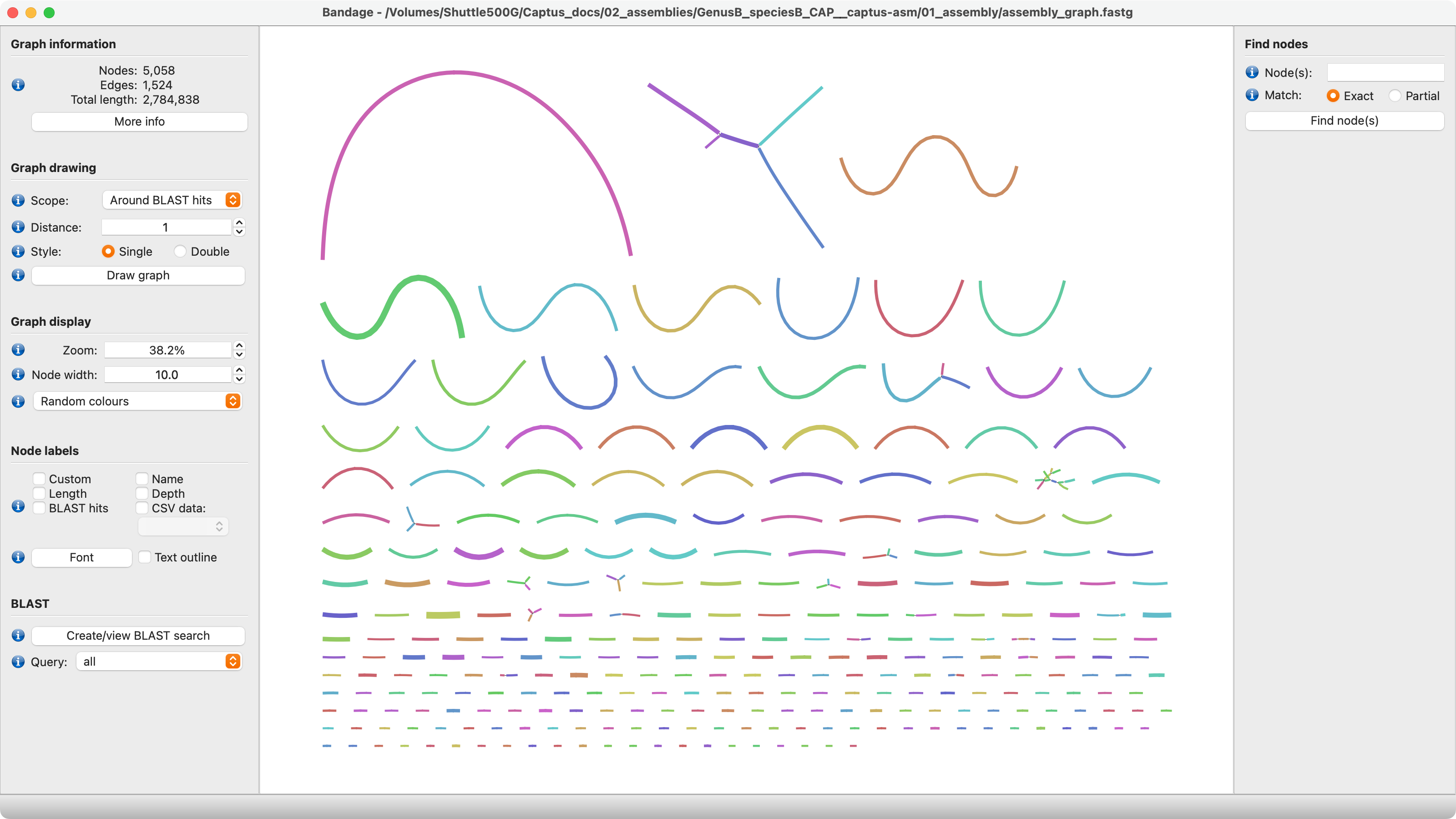
5. assembly_graph.fastg
The assembly graph in FASTG format. This file can be explored in Bandage or similar software which are able to plot the connections between contigs, loops, circular segments, etc. The graph is based on the original MEGAHIT assembly prior to filtering.
6. megahit_brief.log, megahit_full.log
MEGAHIT program logs, the brief version contains just the screen output from each MEGAHIT run.
Example
megahit.brief.log
Captus' MEGAHIT Command:
megahit -1 /Volumes/Shuttle500G/for_docs_output/02_assemblies/GenusA_speciesA_CAP__captus-asm/00_subsampled_reads/GenusA_speciesA_CAP_R1.fq.gz -2 /Volumes/Shuttle500G/for_docs_output/02_assemblies/GenusA_speciesA_CAP__captus-asm/00_subsampled_reads/GenusA_speciesA_CAP_R2.fq.gz --min-count 2 --k-list 31,39,47,63,79,95,111,127,143,159,175 --merge-level 20,0.95 --prune-level 2 --memory 8504035246 --num-cpu-threads 4 --out-dir /Volumes/Shuttle500G/for_docs_output/02_assemblies/GenusA_speciesA_CAP__captus-asm/01_assembly --min-contig-len 182 --tmp-dir /Users/emortiz/captus_megahit_tmp
2022-04-08 01:09:30 - MEGAHIT v1.2.9
2022-04-08 01:09:30 - Using megahit_core with POPCNT and BMI2 support
2022-04-08 01:09:30 - Convert reads to binary library
2022-04-08 01:09:31 - b'INFO sequence/io/sequence_lib.cpp : 77 - Lib 0 (/Volumes/Shuttle500G/for_docs_output/02_assemblies/GenusA_speciesA_CAP__captus-asm/00_subsampled_reads/GenusA_speciesA_CAP_R1.fq.gz,/Volumes/Shuttle500G/for_docs_output/02_assemblies/GenusA_speciesA_CAP__captus-asm/00_subsampled_reads/GenusA_speciesA_CAP_R2.fq.gz): pe, 720090 reads, 151 max length'
2022-04-08 01:09:31 - b'INFO utils/utils.h : 152 - Real: 1.3603\tuser: 0.6603\tsys: 0.2130\tmaxrss: 64000000'
2022-04-08 01:09:31 - Start assembly. Number of CPU threads 4
2022-04-08 01:09:31 - k list: 31,39,47,63,79,95,111,127,143,159,175
2022-04-08 01:09:31 - Memory used: 8504035246
2022-04-08 01:09:31 - Extract solid (k+1)-mers for k = 31
2022-04-08 01:09:41 - Build graph for k = 31
2022-04-08 01:09:43 - Assemble contigs from SdBG for k = 31
2022-04-08 01:09:46 - Local assembly for k = 31
2022-04-08 01:09:55 - Extract iterative edges from k = 31 to 39
2022-04-08 01:09:57 - Build graph for k = 39
2022-04-08 01:09:58 - Assemble contigs from SdBG for k = 39
2022-04-08 01:10:00 - Local assembly for k = 39
2022-04-08 01:10:15 - Extract iterative edges from k = 39 to 47
2022-04-08 01:10:17 - Build graph for k = 47
2022-04-08 01:10:18 - Assemble contigs from SdBG for k = 47
2022-04-08 01:10:20 - Local assembly for k = 47
2022-04-08 01:10:39 - Extract iterative edges from k = 47 to 63
2022-04-08 01:10:41 - Build graph for k = 63
2022-04-08 01:10:42 - Assemble contigs from SdBG for k = 63
2022-04-08 01:10:44 - Local assembly for k = 63
2022-04-08 01:11:03 - Extract iterative edges from k = 63 to 79
2022-04-08 01:11:05 - Build graph for k = 79
2022-04-08 01:11:06 - Assemble contigs from SdBG for k = 79
2022-04-08 01:11:08 - Local assembly for k = 79
2022-04-08 01:11:28 - Extract iterative edges from k = 79 to 95
2022-04-08 01:11:30 - Build graph for k = 95
2022-04-08 01:11:31 - Assemble contigs from SdBG for k = 95
2022-04-08 01:11:33 - Local assembly for k = 95
2022-04-08 01:11:49 - Extract iterative edges from k = 95 to 111
2022-04-08 01:11:50 - Build graph for k = 111
2022-04-08 01:11:51 - Assemble contigs from SdBG for k = 111
2022-04-08 01:11:53 - Local assembly for k = 111
2022-04-08 01:12:08 - Extract iterative edges from k = 111 to 127
2022-04-08 01:12:09 - Build graph for k = 127
2022-04-08 01:12:10 - Assemble contigs from SdBG for k = 127
2022-04-08 01:12:12 - Local assembly for k = 127
2022-04-08 01:12:26 - Extract iterative edges from k = 127 to 143
2022-04-08 01:12:26 - Build graph for k = 143
2022-04-08 01:12:27 - Assemble contigs from SdBG for k = 143
2022-04-08 01:12:29 - Local assembly for k = 143
2022-04-08 01:12:40 - Extract iterative edges from k = 143 to 159
2022-04-08 01:12:40 - Build graph for k = 159
2022-04-08 01:12:41 - Assemble contigs from SdBG for k = 159
2022-04-08 01:12:43 - Local assembly for k = 159
2022-04-08 01:12:53 - Extract iterative edges from k = 159 to 175
2022-04-08 01:12:54 - Build graph for k = 175
2022-04-08 01:12:54 - Assemble contigs from SdBG for k = 175
2022-04-08 01:12:56 - Merging to output final contigs
2022-04-08 01:12:56 - 1850 contigs, total 2031858 bp, min 182 bp, max 26295 bp, avg 1098 bp, N50 1616 bp
2022-04-08 01:12:56 - ALL DONE. Time elapsed: 206.394586 seconds
7. 01_salmon_quant
This directory contains the results of mapping the reads back to the assembled contigs using Salmon. It is not created when --ignore_mapping is used.
8. salmon.log
Salmon logs, combined for the indexing and quantification steps.
9. removed_contigs.fasta
This file is created after the filtering by GC and/or depth is finished (same format as in 4).
10. contigs_depth.tsv
Table containing depth statistics and contig names with the original depth estimated by MEGAHIT and then recalculated with Salmon.
Information included in the table
| Column |
Description |
| megahit_contig_name |
Original contig name from MEGAHIT |
| megahit_depth |
Depth of coverage contained in megahit_contig_name |
| length |
Length of the contig in bp |
| salmon_contig_name |
Contig name with depth of coverage calculated by Salmon |
| salmon_num_reads |
Estimated number of reads mapping to the contig according to Salmon |
| salmon_depth |
read length (multiplied by 2 if reads are paired-end) * salmon_num_reads / length |
11. assembly_stats.tsv
Assembly statistics, before and after filtering:
Information included in the table
| Column |
Description |
| sample_name |
Name of the sample |
| stage |
Before or After filtering |
| num_contigs |
Number of contigs |
| pct_contigs_1kbp |
Percentage of contigs over 1kbp |
| pct_contigs_2kbp |
Percentage of contigs over 2kbp |
| pct_contigs_5kbp |
Percentage of contigs over 5kbp |
| pct_contigs_10kbp |
Percentage of contigs over 10kbp |
| pct_contigs_20kbp |
Percentage of contigs over 20kbp |
| pct_contigs_50kbp |
Percentage of contigs over 50kbp |
| total_length |
Cumulative length of all contigs in bp |
| pct_lengt_1kbp |
Percentage of total assembly length in contigs over 1kbp |
| pct_lengt_2kbp |
Percentage of total assembly length in contigs over 2kbp |
| pct_lengt_5kbp |
Percentage of total assembly length in contigs over 5kbp |
| pct_lengt_10kbp |
Percentage of total assembly length in contigs over 10kbp |
| pct_lengt_20kbp |
Percentage of total assembly length in contigs over 20kbp |
| pct_lengt_50kbp |
Percentage of total assembly length in contigs over 50kbp |
| shortest_contig |
Length of shortest contig in bp |
| longest_contig |
Length of longest contig in bp |
| avg_length |
Average contig length in bp |
| median_length |
Median contig length in bp |
| avg_depth |
Average contig depth |
| median_depth |
Median contig depth |
| gc |
Average contig GC content |
| N50 |
Assembly N50 in bp |
| N75 |
Assembly N75 in bp |
| L50 |
Assembly L50 in number of contigs |
| L75 |
Assembly L75 in number of contigs |
12. depth_stats.tsv
Depth statistics, before and after filtering:
Information included in the table
| Column |
Description |
| sample_name |
Name of the sample |
| stage |
Before or After filtering |
| depth_bin |
Upper limit of the depth bin (lower limit given by the previous depth bin value) |
| length |
Sum of lengths of the contigs inside the depth_bin |
| fraction |
Sum of lengths of the contigs inside the depth_bin as a fraction of the total length |
| num_contigs |
Number of contigs inside the depth_bin |
13. length_stats.tsv
Length statistics, before and after filtering:
Information included in the table
| Column |
Description |
| sample_name |
Name of the sample |
| stage |
Before or After filtering |
| length_bin |
Upper limit of the length bin (lower limit given by the previous length bin value) |
| length |
Sum of lengths of the contigs inside the length_bin |
| fraction |
Sum of lengths of the contigs inside the length_bin as a fraction of the total length |
| num_contigs |
Number of contigs inside the length_bin |
14. captus-assemble_assembly_stats.tsv
Assembly statistics compiled across all samples, before and after filtering:
Information included in the table
| Column |
Description |
| sample_name |
Name of the sample |
| stage |
Before or After filtering |
| num_contigs |
Number of contigs |
| pct_contigs_1kbp |
Percentage of contigs over 1kbp |
| pct_contigs_2kbp |
Percentage of contigs over 2kbp |
| pct_contigs_5kbp |
Percentage of contigs over 5kbp |
| pct_contigs_10kbp |
Percentage of contigs over 10kbp |
| pct_contigs_20kbp |
Percentage of contigs over 20kbp |
| pct_contigs_50kbp |
Percentage of contigs over 50kbp |
| total_length |
Cumulative length of all contigs in bp |
| pct_lengt_1kbp |
Percentage of total assembly length in contigs over 1kbp |
| pct_lengt_2kbp |
Percentage of total assembly length in contigs over 2kbp |
| pct_lengt_5kbp |
Percentage of total assembly length in contigs over 5kbp |
| pct_lengt_10kbp |
Percentage of total assembly length in contigs over 10kbp |
| pct_lengt_20kbp |
Percentage of total assembly length in contigs over 20kbp |
| pct_lengt_50kbp |
Percentage of total assembly length in contigs over 50kbp |
| shortest_contig |
Length of shortest contig in bp |
| longest_contig |
Length of longest contig in bp |
| avg_length |
Average contig length in bp |
| median_length |
Median contig length in bp |
| avg_depth |
Average contig depth |
| median_depth |
Median contig depth |
| gc |
Average contig GC content |
| N50 |
Assembly N50 in bp |
| N75 |
Assembly N75 in bp |
| L50 |
Assembly L50 in number of contigs |
| L75 |
Assembly L75 in number of contigs |
15. captus-assemble_depth_stats.tsv
Depth statistics compiled across all samples, before and after filtering:
Information included in the table
| Column |
Description |
| sample_name |
Name of the sample |
| stage |
Before or After filtering |
| depth_bin |
Upper limit of the depth bin (lower limit given by the previous depth bin value) |
| length |
Sum of lengths of the contigs inside the depth_bin |
| fraction |
Sum of lengths of the contigs inside the depth_bin as a fraction of the total length |
| num_contigs |
Number of contigs inside the depth_bin |
16. captus-assemble_length_stats.tsv
Length statistics compiled across all samples, before and after filtering:
Information included in the table
| Column |
Description |
| sample_name |
Name of the sample |
| stage |
Before or After filtering |
| length_bin |
Upper limit of the length bin (lower limit given by the previous length bin value) |
| length |
Sum of lengths of the contigs inside the length_bin |
| fraction |
Sum of lengths of the contigs inside the length_bin as a fraction of the total length |
| num_contigs |
Number of contigs inside the length_bin |
17. captus-assemble_report.html
This is the final Assembly report, summarizing statistics across all samples assembled.
18. captus-assemble.log
This is the log from Captus, it contains the command used and all the information shown during the run. If the option --show_less was enabled, the log will also contain all the extra detailed information that was hidden during the run.
Created by Edgardo M. Ortiz (06.08.2021)
Last modified by Edgardo M. Ortiz (23.12.2024)