Input Preparation
Before starting your analysis, a VERY IMPORTANT step is to rename your FASTQ files so they clearly identify your samples throughout the entire analysis.
In general, a good tip for renaming your samples is to think on how you want the names in your final phylogenetic tree.
The only special characters that are safe to use in the sample name are -, and _ (_ is commonly used to replace spaces in many phylogenetic programs). Otherwise, do not use spaces, other special characters (! " # $ % & ( ) * + , . / : ; < = > ? @ [ \ ] ^ ` { | } ~), or accented letters (like á, è, ü, ç, ñ), they are just guaranteed to give you headaches at some point.
Also, please use this naming convention for your FASTQ files:
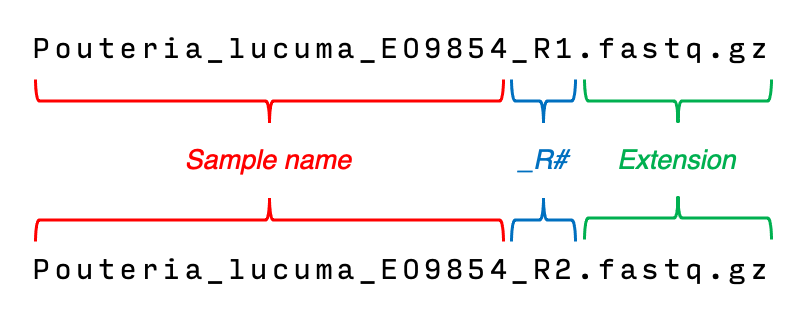
- IMPORTANT: Even though underscores (
_) are allowed in sample names, please DO NOT use more than ONE consecutive_in any case. We use double underscores (__) internally to separate several pieces of information during the processing and for the output (see for example the FASTA headers of extracted markers). - Any text found before the _R# pattern and the extension will become your sample name (
Pouteria_lucuma_EO9854in this case). - If you are using paired-end reads, your R1 and R2 filenames should contain the patterns
_R1and_R2respectively to be correctly matched and used as pairs. For single-end your filenames should still contain_R1. - These are the valid extensions:
.fq,.fastq,.fq.gz, and.fastq.gz.
These are examples of valid FASTQ filenames for Captus:
Arabidopsis_thaliana_R1.fq.gzandArabidopsis_thaliana_R2.fq.gz, these will be correctly taken as a pairMus_musculus_GX763763_R1.fastq, if its corresponding R2 is not found it will be used as single-endERI_Demosthenesia_mandonii_EO2765_R1.fastq.gzandERI_Demosthenesia_mandonii_EO2765_R2.fastq.gzA_R1.fqandA_R2.fq
And here, some examples or invalid FASTQ filenames:
ERR246535_1.fastq.gzandERR246535_2.fastq.gz, notice they lack the_R1and_R2patterns in the names,Captusis not able to match these as a pairOctomeles_sp.1_R1.fastq, it is better to replace the.in the sample name by a-to getOctomeles_sp-1_R1.fastqMalus_doméstica.fast, the sample name contains and accenté, but most importantly, it will be ignored because of the invalid extension.fast
Created by Edgardo M. Ortiz (06.08.2021)
Last modified by Edgardo M. Ortiz (30.05.2022)